Advanced R for Econometricians
Improving Performance
Martin C. Arnold
Improving Performance
 # packages neededlibrary(tidyverse) # for bench packagelibrary(parallel)library(MonteCarlo)
# packages neededlibrary(tidyverse) # for bench packagelibrary(parallel)library(MonteCarlo)We should forget about small efficiencies, say about 97% of the time: premature optimisation is the root of all evil. Yet we should not pass up our opportunities in that critical 3%. A good programmer will not be lulled into complacency by such reasoning, he will be wise to look carefully at the critical code; but only after that code has been identified. — Donald Knuth
Improving Performance — Overview
Once profiling has revealed a bottleneck, we need to make it faster. In the following we will discuss some techniques that are broadly useful:
Look for existing solutions.
Searching google can save you lots of time. The benefit of finding a faster (but possibly suboptimal) solution on your own may be small.
Do as little as possible. Be precise.
Using optimized functions tailored for specific tasks often makes a big difference.
Vectorise your code.
This often relates to avoiding loops and the associated inefficiencies.
Parallelise (if possible).
Using multiple CPU cores in parallel is enormously helpful when the computation involves a large number of tasks that may be solved independently.
1. Look for Existing Solutions
There's a good chance that someone has already tackled the same problem.
Find out if there is a Cran Task View related to your problem and search the packages listed there
Limit your search to packages that depend on the
Rcpppackage (and thus likely implement high-performant C++ code). This is easily done by checking reverse dependencies of Rcpp.Check if a question related to your problem has been asked on stackoverflow. Narrow the search by using suitable tags, e.g.,
[r],[performance].Google. For R-related results, use rseek.
Record all solutions that you find not just those that immediately appear to be faster!
Some solutions might be initially slower but are are easier to optimize and thus end up being faster.
Sometimes combining the fastest parts from different approaches is helpful.
Improving Performance — Golden Rule
Before we continue, we introduce a rule we have already followed implicitly: the golden rule of R programming:
Access the underlying C/FORTRAN routines as quickly as possible; the fewer functions calls required to achieve this, the better.
(Lovelace and Gillespie, 2016)

The techniques discussed next follow this paradigm. If this is not enough, we still may rewrite (some of) the code in C++ using the Rcpp package (discussed in the next chapter).
2. Do as Little as Possible
If you cannot find existing solutions, start by reducing your code to the bare essentials.
Exercise: coercion of inputs / robustness checks
 X <- matrix(1:1000, ncol = 10)Y <- as.data.frame(X)
X <- matrix(1:1000, ncol = 10)Y <- as.data.frame(X)Which approach is faster and why?
 apply(X, 1, sum)apply(Y, 1, sum)
apply(X, 1, sum)apply(Y, 1, sum)2. Do as Little as Possible
A function is faster if it has less to do — obvious but often neglected!
Use functions tailored to specific input and output types / specific problems!
Exercise: coercion of inputs / robustness checks — ctd.
Which approach is faster and why?
 rowSums(X)apply(X, 1, sum)
rowSums(X)apply(X, 1, sum)2. Do as Little as Possible
Exercise: searching a vector
What is the fastest way to check if 10 is an element of 1:100?
2. Do as Little as Possible
Exercise: Linear Regression — computation of SE(ˆβ)
Which approach is faster and why? Name disadvantages of the fastest one.
 set.seed(1)X <- matrix(rnorm(100), ncol = 1)Y <- X + rnorm(100)a <- function() { coefficients(summary(lm(Y ~ X - 1)))[1, 2] }b <- function() { fit <- lm.fit(X, Y) c( sqrt(1/(length(X)-1) * sum(fit$residuals^2) * solve(t(X) %*% X)) )}
set.seed(1)X <- matrix(rnorm(100), ncol = 1)Y <- X + rnorm(100)a <- function() { coefficients(summary(lm(Y ~ X - 1)))[1, 2] }b <- function() { fit <- lm.fit(X, Y) c( sqrt(1/(length(X)-1) * sum(fit$residuals^2) * solve(t(X) %*% X)) )}2. Do as Little as Possible
Be as explicit as possible.
Example: method dispatch takes time
 x <- runif(1e2)bench::mark( mean(x), mean.default(x))
x <- runif(1e2)bench::mark( mean(x), mean.default(x))## # A tibble: 2 x 6## expression min median `itr/sec` mem_alloc `gc/sec`## <bch:expr> <bch:tm> <bch:tm> <dbl> <bch:byt> <dbl>## 1 mean(x) 2.64µs 3.07µs 297320. 22.7KB 0 ## 2 mean.default(x) 1.13µs 1.35µs 659992. 0B 66.02. Do as Little as Possible
Benchmark against alternatives that rely on primitive functions.
Example: primitives are faster
 bench::mark( mean.default(x), sum(x)/length(x))
bench::mark( mean.default(x), sum(x)/length(x))## # A tibble: 2 x 6## expression min median `itr/sec` mem_alloc `gc/sec`## <bch:expr> <bch:tm> <bch:tm> <dbl> <bch:byt> <dbl>## 1 mean.default(x) 1.15µs 1.38µs 658724. 0B 0## 2 sum(x)/length(x) 432ns 526ns 1637111. 0B 02. Do as Little as Possible — Exercises
- Can you come up with an even faster implementation of
b()in the linear regression example?
What’s the difference between
rowSums()and.rowSums()?rowSums2()is an alternative implementation ofrowSums(). Is it faster for the inputdf? Why?rowSums2 <- function(df) {out <- df[[1L]]if (ncol(df) == 1) return(out)for (i in 2:ncol(df)) {out <- out + df[[i]]}out}df <- as.data.frame(replicate(1e3, sample(100, 1e4, replace = TRUE)))
2. Do as Little as Possible — Case Study
Imagine you want to compute the bootstrap distribution of a sample correlation coefficient using cor_df() and the data in the example below.
Given that you want to run this many times, how can you make this code faster?
 n <- 1e6df <- data.frame(a = rnorm(n), b = rnorm(n))cor_df <- function(df, n) { i <- sample(seq(n), n, replace = TRUE) cor(df[i, , drop = FALSE])[2, 1]}
n <- 1e6df <- data.frame(a = rnorm(n), b = rnorm(n))cor_df <- function(df, n) { i <- sample(seq(n), n, replace = TRUE) cor(df[i, , drop = FALSE])[2, 1]}3. Vectorise your code
What is vectorisation?
Vectorisation is the process of converting an algorithm from operating on a single value at a time to operating on a set of values (like a vector) at one time.
It makes problems simpler: code that operates on vectors instead of single entries of an array is often less complex to write.
Why is it efficient?
(on the lowest level where functions map closely to processor instructions)
- Your computer is designed to run vectorised operations: a vectorised function may run multiple operations from a single instruction which is faster than sending individual instructions for each operation.
3. Vectorise your code
Why is vectorisation efficient? — ctd.
Many languages (including R) work on arrays that are stored in the memory in column-major order.
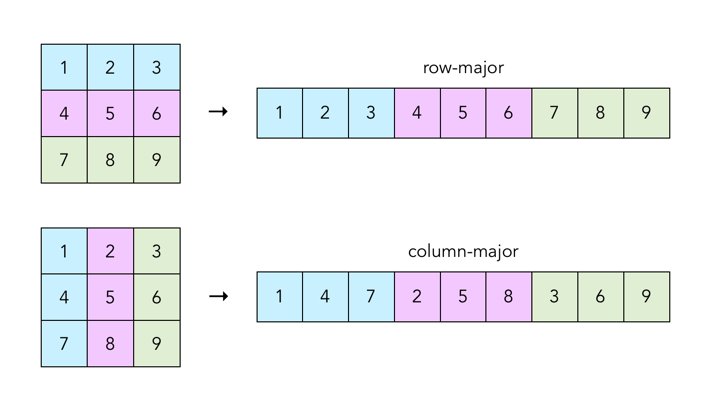
Disregarding these patterns by writing code which operates on 1×1 vectors thus means 'fighting the language'.
3. Vectorise your code
Why is vectorisation efficient? — ctd.
Example: vector addition
⎛⎜⎝123⎞⎟⎠+⎛⎜⎝456⎞⎟⎠=⎛⎜⎝579⎞⎟⎠
Slow: three instructions, three operations
- add 1 and 4
- add 2 and 5
- add 3 and 6
Fast: one instruction, one vectorised operation:
add (1 2 3)′ and (4 5 6)′
The three additions are effectively done in parallel.
Aagain, less related to R itself but to what the CPU does.
3. Vectorise your code
What does it mean to write 'vectorised' code in R?
Remember that there are no 'real' scalars in R. Everything that looks like a scalar is actually a 1×1 vector.
 # otherwise this shouldn't work:1[1]
# otherwise this shouldn't work:1[1]## [1] 1Thus 'scalar' operations work on vector elements which is (often) needlessly cumbersome.
3. Vectorise your code
What does it mean to write 'vectorised' code in R?
By 'vectorised' we mean that the function works on vectors, i.e., performs vector arithmetic and calls functions which work on vectors.
Example: scalar vs. vectorised computation of euclidean norm
 L2_scalar <- function(x) { out <- numeric(1) for(i in 1:length(x)) { out <- x[i] * x[i] + out } return(sqrt(out))}
L2_scalar <- function(x) { out <- numeric(1) for(i in 1:length(x)) { out <- x[i] * x[i] + out } return(sqrt(out))} L2_vec <- function(x) { return( sqrt( sum(x * x) ) )}
L2_vec <- function(x) { return( sqrt( sum(x * x) ) )}3. Vectorise your code
What does it mean to write 'vectorised' code in R?
By 'vectorised' we mean that the function works on vectors, i.e., performs vector arithmetic and calls functions which work on vectors.
Example: scalar vs. vectorised computation of euclidean norm
 bench::mark( L2_scalar(1:1e4), L2_vec(1:1e4))
bench::mark( L2_scalar(1:1e4), L2_vec(1:1e4))## # A tibble: 2 x 6## expression min median `itr/sec` mem_alloc `gc/sec`## <bch:expr> <bch:tm> <bch:tm> <dbl> <bch:byt> <dbl>## 1 L2_scalar(1:10000) 713.1µs 775.8µs 1236. 4.11MB 0 ## 2 L2_vec(1:10000) 40.3µs 73.1µs 12708. 78.22KB 16.93. Vectorise your code — Some Principles
Do not torture the interpreter
Vectorisation reduces the amount of intepreting R has to do: Validating that
x <- 1Landy <- 1:100Lare of typeintegeris equally expensive. Checking that each element ofyis an integer is not!Use functions from base R that work on vectors
Vectorisation may avoid loops in R. Loops in many 'vectorised' R functions are carried out in C and thus are much faster.
The following functions are prominent examples:
rowSums(),colSums(),rowMeans(),colMeans(),cumsum(),diff()Use matrix algebra
Most matrix operations that involve loops are executed by highly optimized external Fortran and C libraries. See, e.g., BLAS.
3. Vectorise your code
Are for() loops slower than *apply()?
for() loops have the reputation of being slow(er than *apply()). This is widespread wisdom and not generally true:
for()is not slow if we iterate over data and apply a (non-vectorised) function. Execution time is comparable to*apply().Loops are often used in an inefficient manner:
Growing an object and improper initialisation means overhead due to growing / copying in every iteration.
Getting used to
*apply()is a good practice as it prevents us from doing silly stuff like the above.
What if I need to use for / how to properly write a for loop?
Allocate the required storage before the loop and fill the object!
3. Vectorise your code
Example: avoid growing objects
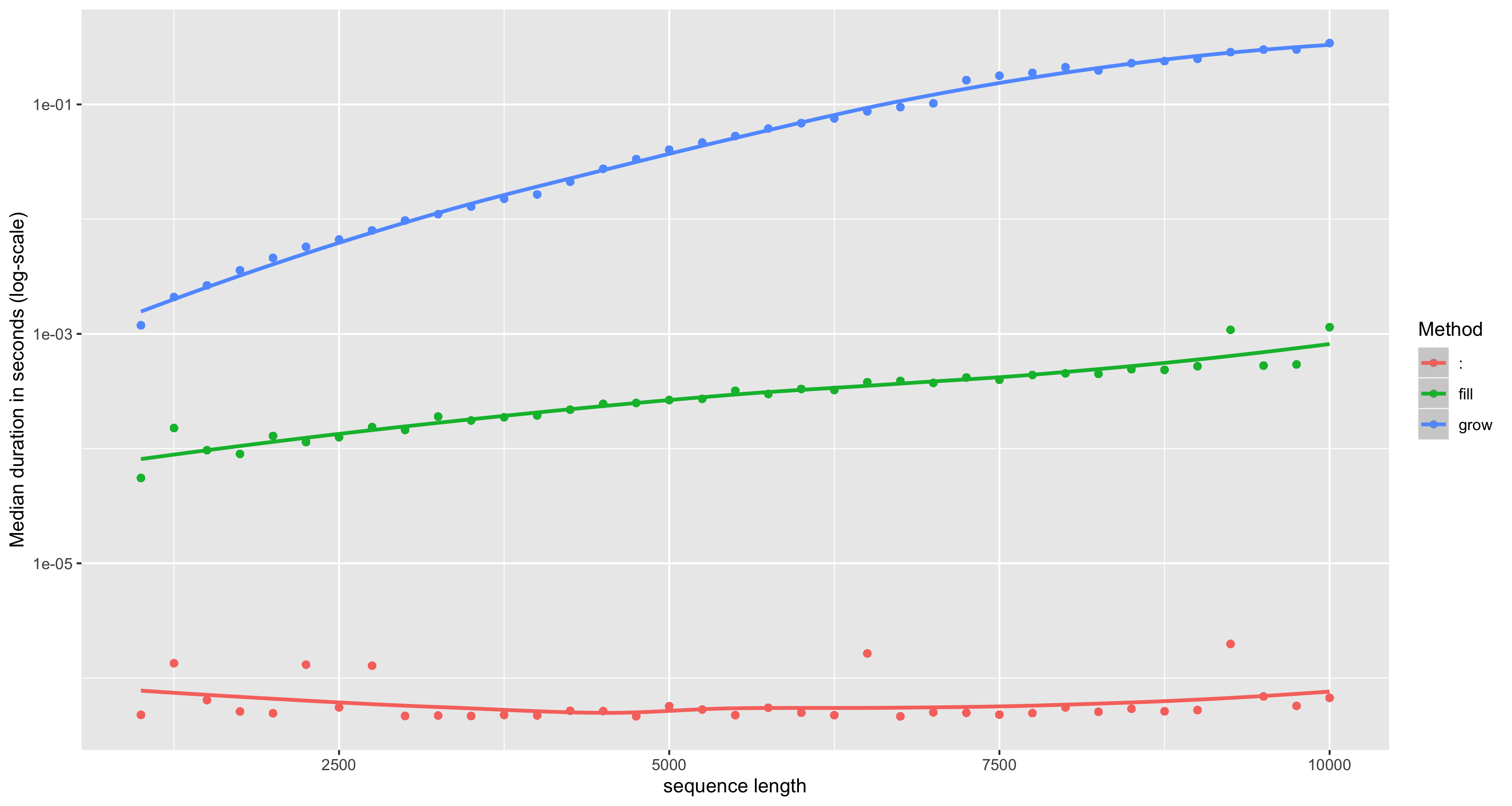
3. Vectorise your code
Example: good and bad for() loops
 # Bad: c()/cbind()/rbind()rw_bad <- function(N) { set.seed(1) out <- rnorm(1) for(i in 2:N) { out <- c(out, out[i-1] + rnorm(1)) } return(out)}
# Bad: c()/cbind()/rbind()rw_bad <- function(N) { set.seed(1) out <- rnorm(1) for(i in 2:N) { out <- c(out, out[i-1] + rnorm(1)) } return(out)}
3. Vectorise your code
Example: good and bad for() loops
 # Good: proper initialisation/iterationrw_good <- function(N) { set.seed(1) out <- vector("double", N) out[1] <- rnorm(1) for(i in 2:N) { out[i] <- rnorm(1) + out[i-1] } return(out)}
# Good: proper initialisation/iterationrw_good <- function(N) { set.seed(1) out <- vector("double", N) out[1] <- rnorm(1) for(i in 2:N) { out[i] <- rnorm(1) + out[i-1] } return(out)}
3. Vectorise your code
Example: good and bad for() loops
 bench::mark(rw_good(1e4), rw_bad(1e4))
bench::mark(rw_good(1e4), rw_bad(1e4))## # A tibble: 2 x 6## expression min median `itr/sec` mem_alloc `gc/sec`## <bch:expr> <bch:tm> <bch:tm> <dbl> <bch:byt> <dbl>## 1 rw_good(10000) 15.2ms 22.6ms 39.3 24.8MB 21.6## 2 rw_bad(10000) 346ms 351.5ms 2.85 406.3MB 27.03. Vectorise your code
Are for() loops slower than the *apply() functions?
Example: for() vs. apply()
 X <- matrix(rnorm(1e4), ncol = 1e4)colmax <- function(x) { out <- numeric(ncol(x)) for(j in 1:ncol(x)) { out[j] <- max(x[,j]) } return(out)}
X <- matrix(rnorm(1e4), ncol = 1e4)colmax <- function(x) { out <- numeric(ncol(x)) for(j in 1:ncol(x)) { out[j] <- max(x[,j]) } return(out)}3. Vectorise your code
Are for() loops slower than the *apply() functions?
Example: for() vs. apply() — ctd.
 bench::mark( colmax(X), apply(X, 2, max))
bench::mark( colmax(X), apply(X, 2, max))## # A tibble: 2 x 6## expression min median `itr/sec` mem_alloc `gc/sec`## <bch:expr> <bch:tm> <bch:tm> <dbl> <bch:byt> <dbl>## 1 colmax(X) 5.75ms 6.37ms 156. 4.34MB 55.0## 2 apply(X, 2, max) 9.05ms 10.27ms 96.6 312.73KB 55.23. Vectorise your code — Exercises
Compare the speed of
apply(X, 1, sum)with the vectorisedrowSums(X)for varying sizes of the square matrixXusingbench::mark(). Consider the dimensions1,1e1,1e2,1e3,0.5e4and1e5. Visualize the results using a violin plot.(a) How can you vectorise the computation of a weighted sum?
(b) How can you use
crossprod()to compute that same sum?(c) Compare performance of the approaches in (a) and (b)
Find an approach to compute column maxima of a numeric matrix
Xusing a*apply()-function which outperformsapply(X, 2, max).
3. Vectorise your code — Case Study
Case Study: Monte Carlo Integration
Suppose you are interested in computing A=∫10x2dx using Monte Carlo (MC) integration. Consider the following algorithm which operates on 1×1 vectors in a for() loop:
initialize
countsfor i in 1:NGenerate Ui∼i.i.d.U(0,1); i=1,2.
If U2<U21, then
counts = counts + 1end
forEstimate A by
counts/N
3. Vectorise your code — Case Study
Case Study: Monte Carlo Integration
(a) Implement the algorithm in a function
A_loop(N). A should use a loop to compute the MC estimate of A withNMC iterations.(b) Estimate A using
A_loop(N)with 5e5 MC iterations.(a) Define a function
A_vec(N)which implements the MC algorithm using a vectorised approach.(b) Compare
A_vec(N)andA_loop(N)in a microbenchmark forN = 5e5iterations.(c) How does the difference in performance relate to
N?
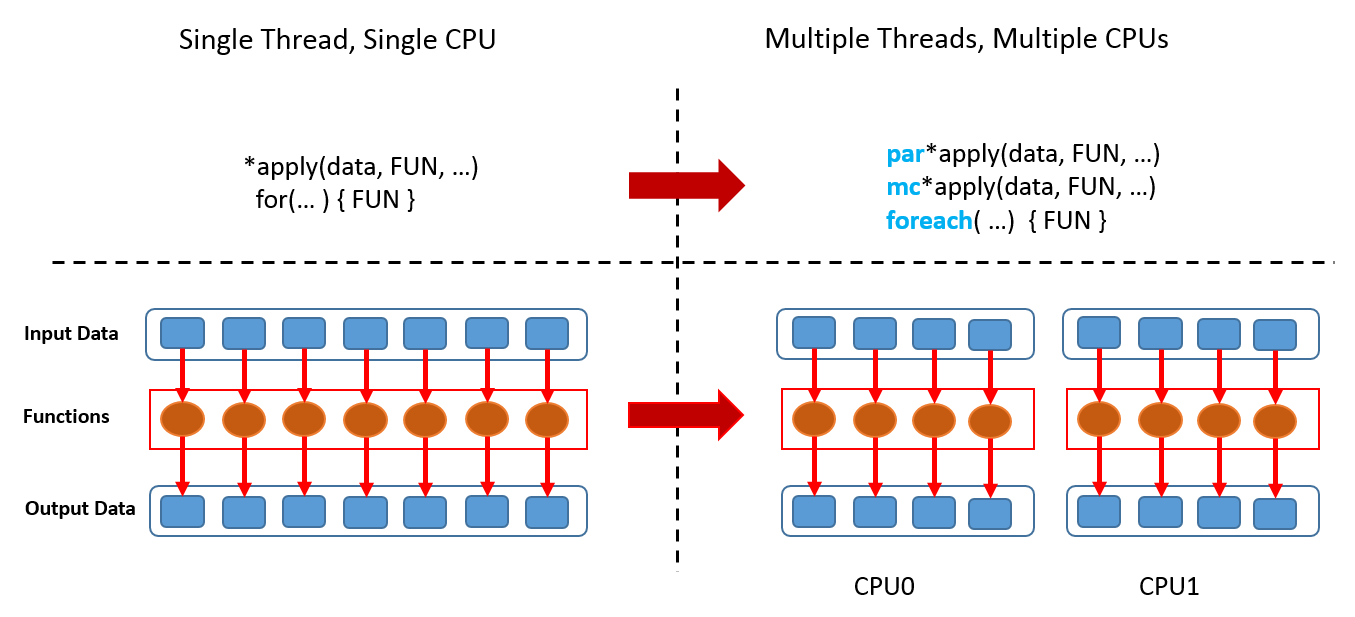
4. Parallelisation — FAQs
What is parallelisation?
In the simplest sense, parallelisation means the simultaneous use of multiple hardware resources for solving a computational problem.
In the following we refer to parallelisation as the distribution of computing tasks over several CPUs in a shared memory system (a system where multiple CPUs access the same memory).
Does R perform parallelisation by default?
No. The 'stock' R version on CRAN is a single-threaded program, i.e., R does not benefit from modern multi-threaded CPUs, multiple cores let alone multiple CPUs.
4. Parallelisation — FAQs
Is it complicated to parallelise code?
In general, yes. Fortunately, there are R packages for the most common platforms that do the work for us. These packages facilitate the process substantially.
In most cases, we don't even have to adjust our code in order for it to be executable in parallel.
Which tasks can be done in parallel?
In principle, all operations that can be executed independently.
Parallel computing is often used in MC simulations which require computation of a large number of homogeneous and independent tasks.
Is parallelisation always faster than 'serial' computation?
No. The cost of managing the computations (the overhead) may offset or even surpass time savings gained by parallel computation. This occurs, e.g., when a small number of simple tasks is solved in parallel. We will discuss an example.
4. Parallelisation — the parallel package
The parallel package (comes with base R) is a good starting point for parallel computing.
 # detect numer of cores (including logical cores)library(parallel)detectCores()
# detect numer of cores (including logical cores)library(parallel)detectCores()## [1] 8The notebook used to run this script has a 2,3 GHz Intel Core i5 processor with 4 physical CPUs and supports hyper-threading, i.e., each CPU has 2 logical cores.
 # detect numer of physical coresdetectCores(logical = F)
# detect numer of physical coresdetectCores(logical = F)## [1] 4Note that the behavior of detectCores() is platform specific and not always reliable, see ?detectCores().
4. Parallelisation - parallel::mclapply()
Example: send them cores to sleep (on MacOS)
 r <- mclapply(1:8, function(i) Sys.sleep(20), mc.cores = 8)
r <- mclapply(1:8, function(i) Sys.sleep(20), mc.cores = 8)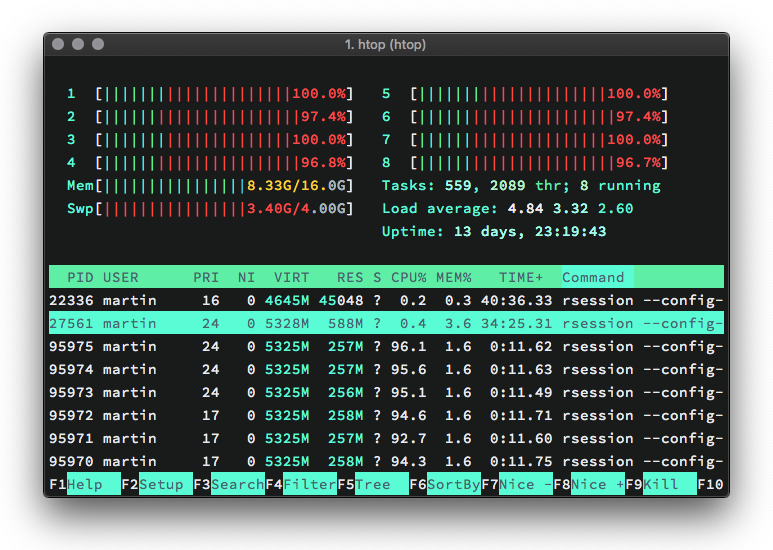
4. Parallelisation - parallel::mclapply()
Example: send them cores to sleep (on MacOS) — ctd.
What happened?
The R session (the main Process) opened 8 sub-processes, so-called forks (labeled
rsessionin the activity monitor)The forks operate independently from the main process (but they 'know' about each other). Each fork operates on a separate core.
mclapply()passes a call offunction(i) Sys.sleep(20)to each fork. Computation (or rather doing nothing for 20 seconds) now proceeds in parallel.After 20 seconds the results are gathered in
rand all forks are killed. Note thatris just a list withNULLentries.
4. Parallelisation - parallel::mclapply()
Example: send them cores to sleep (Windows)
An alternative on Windows is to use parallel::parLapply(). The setup is somewhat more complex.
 # set number of cores to be usedcores <- detectCores(logical = TRUE) - 1# initialize clustercl <- makeCluster(cores)# run lapply in parallelparLapply(cl, 1:8, function(i) Sys.sleep(20))# stop clusterstopCluster(cl)
# set number of cores to be usedcores <- detectCores(logical = TRUE) - 1# initialize clustercl <- makeCluster(cores)# run lapply in parallelparLapply(cl, 1:8, function(i) Sys.sleep(20))# stop clusterstopCluster(cl)4. Parallelisation
Example: parallelised bootstrapping
Remember the function cor_df() from the case study in 2. Do as Little as Possible. A faster approach, cor_df2(), is given below.
 n <- 1e4df <- data.frame(a = rnorm(n), b = rnorm(n))cor_df2 <- function(x, n) { i <- sample.int(n, n, replace = T) cor(df$a[i], df$b[i])}
n <- 1e4df <- data.frame(a = rnorm(n), b = rnorm(n))cor_df2 <- function(x, n) { i <- sample.int(n, n, replace = T) cor(df$a[i], df$b[i])}cor_df2() would be even faster if df was a matrix object.
function(df, n) { i <- sample(1:n, n, replace = T) m <- df[i,] cor(m)[2, 1]}4. Parallelisation
Example: parallelised bootstrapping — ctd.
Calling cor_df2() a large number of times is time consuming.
 # serial computations_ser <- system.time( r_ser <- lapply(1:1e5, function(x) cor_df2(df, n)))s_ser
# serial computations_ser <- system.time( r_ser <- lapply(1:1e5, function(x) cor_df2(df, n)))s_ser## ## user system elapsed ## 41.258 7.497 48.8354. Parallelisation
Example: parallelised bootstrapping — ctd.
Luckily, it is straightforward to bootstrap in parallel using mclapply().
 # parallel computations_par <- system.time( r_par <- mclapply(1:1e5, function(x) cor_df2(df, n), mc.cores = 8))s_par
# parallel computations_par <- system.time( r_par <- mclapply(1:1e5, function(x) cor_df2(df, n), mc.cores = 8))s_par## ## user system elapsed ## 38.343 7.995 16.343Note that using 8 cores in my case is (only) ~5 times faster than the serial approach due to overhead.
4. Parallelisation
Example: too much overhead
Parallelisation may be inefficient:
 system.time(mclapply(1:1e5, sqrt, mc.cores = 8))
system.time(mclapply(1:1e5, sqrt, mc.cores = 8))## ## ## user system elapsed ## ## 0.107 0.110 0.062 ## system.time(lapply(1:1e5, sqrt))
system.time(lapply(1:1e5, sqrt))## ## ## user system elapsed ## ## 0.034 0.001 0.035 ##This is because the cost of the individual computations is low, and additional work is needed to send the computation to the different cores and to collect the results: The overhead exceeds the time savings.
4. The MonteCarlo Package
Why is worthwhile to get used to such packages?
Running MC simulations is an everyday task in econometrics: simulations are routinely used for approximating quantities which cannot be derived analytically, e.g., finite sample properties of a test statistic.
MC simulation is also an essential tool in statistical programming because we often write functions that expect random input and/or produce random output
Testing whether the impementation is correct amounts to checking if the statistical properties of the output are as expected, using a large number of samples
Often, the effort to write the simulation exceeds the effort to implement the function by a multiple
4. The MonteCarlo Package
The MonteCarlo package streamlines the process of writing MC simulations.
Useful features are:
Simulation over a user-defined parameter grid
Automatic parallelisation using the
snowpackageCompilation of the results in an array. Generation of customisable LaTeX tables is possible.
The package is available on CRAN.
Development version @ https://github.com/FunWithR/MonteCarlo
4. The MonteCarlo Package
Example: Unit Root test
Consider the DGP yt=ρyt−1+ϵt, ϵt∼N(0,1) with y0=0 and ρ=1. It is a well known result that the limit of the t-ratio t=^ρ−1SE(^ρ) can be written as a functional of Wiener processes,
t⇒[W(1)]2−12√∫10[W(r)]2dr.
The asymptotic distribution of t is non-normal and its quantiles cannot be derived analytically.
4. The MonteCarlo Package
Example: Unit Root test — ctd.
We use the MonteCarlo package to simulate the limit distribution of t.
 library(MonteCarlo)
library(MonteCarlo) sim <- function(rho, Time) { Y <- matrix(NA_real_, ncol = 1, nrow = Time); Y[1,1] <- rnorm(1) for(t in 2:Time) { Y[t, 1] <- rho * Y[t-1, 1] + rnorm(1) } y <- Y[-1, , drop = FALSE] ylag <- Y[-Time, , drop = FALSE] fit <- lm.fit(ylag, y) rho <- fit$coefficients res <- fit$residuals sigma <- sqrt(1/(Time-2) * t(res) %*% res * 1/ (t(ylag) %*% ylag)) return( list("t" = (rho[[1]] - 1)/sigma) )}
sim <- function(rho, Time) { Y <- matrix(NA_real_, ncol = 1, nrow = Time); Y[1,1] <- rnorm(1) for(t in 2:Time) { Y[t, 1] <- rho * Y[t-1, 1] + rnorm(1) } y <- Y[-1, , drop = FALSE] ylag <- Y[-Time, , drop = FALSE] fit <- lm.fit(ylag, y) rho <- fit$coefficients res <- fit$residuals sigma <- sqrt(1/(Time-2) * t(res) %*% res * 1/ (t(ylag) %*% ylag)) return( list("t" = (rho[[1]] - 1)/sigma) )}4. The MonteCarlo Package
Example: Unit Root test — ctd.
 # setup list of parametersparams <- list("rho" = 1, "Time" = 1000)# run parallelised MC simulation over parameter gridr <- MonteCarlo( sim, nrep = 50000, param_list = params, ncpus = parallel::detectCores())
# setup list of parametersparams <- list("rho" = 1, "Time" = 1000)# run parallelised MC simulation over parameter gridr <- MonteCarlo( sim, nrep = 50000, param_list = params, ncpus = parallel::detectCores())We transform the simulated test statistics to a vector.
 t_sim <- apply(r$result$t, 3, c)
t_sim <- apply(r$result$t, 3, c)4. The MonteCarlo Package
Example: Unit Root test — ctd.
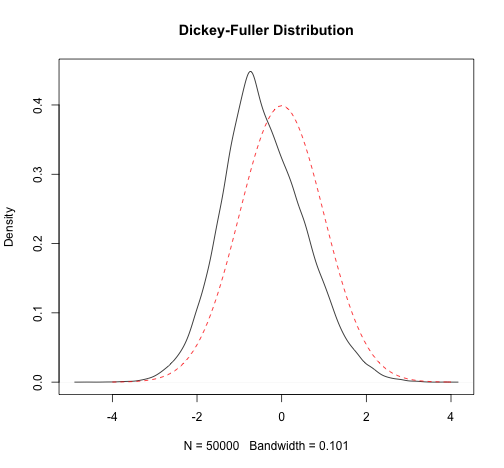
Estimates of the finite sample quantiles are close to the quantiles of the corresponding asymptotic Dickey-Fuller distribution
 quantile(t_sim, c(0.05, 0.1, 0.5, 0.9, 0.95))
quantile(t_sim, c(0.05, 0.1, 0.5, 0.9, 0.95))## 5% 10% 50% 90% 95% ## -1.9472738 -1.6124977 -0.5044926 0.8846489 1.2702348 fUnitRoots::qunitroot(c(0.05, 0.1, 0.5, 0.9, 0.95), trend = "nc")
fUnitRoots::qunitroot(c(0.05, 0.1, 0.5, 0.9, 0.95), trend = "nc")## [1] -1.9408468 -1.6167526 -0.4999276 0.8877673 1.2836012We compare with the Gaussian distribution.
 plot(density(t_sim), main = "Dickey-Fuller Distribution")curve(dnorm(x), from = -4, to = 4, add = T, lty = 2, col = "red")
plot(density(t_sim), main = "Dickey-Fuller Distribution")curve(dnorm(x), from = -4, to = 4, add = T, lty = 2, col = "red")4. The MonteCarlo Package
Example: Unit Root test — ctd.
4. The MonteCarlo Package
It is a great benefit to work with the package if you want to run the simulation for several parameter combinations.
Example: Power of DF-Test
We write a simple wrapper for sim() which returns a logical value for rejection at 5% using the asymptotic critical value.
 # asymptotic 5% critical value of the corresponding DF-distributioncrit <- fUnitRoots::qunitroot(0.05, trend = "nc")# rejection?sim_pow <- function(rho, Time) { return( list("t" = sim(rho, Time)[[1]] < crit) )}
# asymptotic 5% critical value of the corresponding DF-distributioncrit <- fUnitRoots::qunitroot(0.05, trend = "nc")# rejection?sim_pow <- function(rho, Time) { return( list("t" = sim(rho, Time)[[1]] < crit) )}4. The MonteCarlo Package
Example: Power of DF-Test — ctd.
We consider all combination for a sequence of alternatives and three different sample sizes.
 # setup list of parameters (yielding 33 parameter constellations)params <- list("rho" = seq(0.75, 1, 0.025), "Time" = c(50, 100, 200))# run parallelised MC simulation over parameter gridr <- MonteCarlo( sim_pow, nrep = 5000, param_list = params, ncpus = parallel::detectCores())
# setup list of parameters (yielding 33 parameter constellations)params <- list("rho" = seq(0.75, 1, 0.025), "Time" = c(50, 100, 200))# run parallelised MC simulation over parameter gridr <- MonteCarlo( sim_pow, nrep = 5000, param_list = params, ncpus = parallel::detectCores())4. The MonteCarlo Package
Example: Power of DF-Test — ctd.
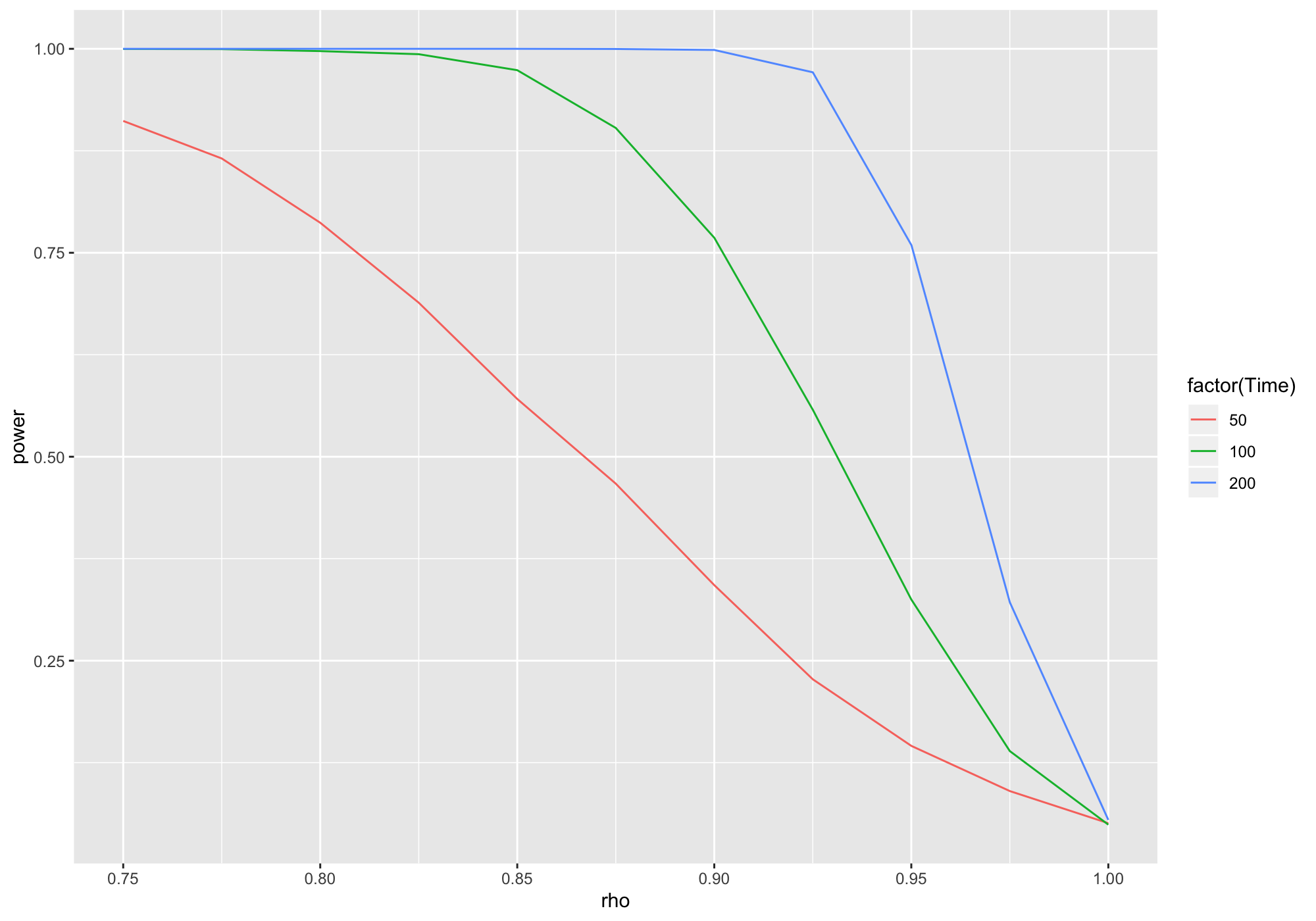
Content of a MonteCarlo object is easily transformed and visualised using tidyverse functions.
 library(dplyr)library(ggplot2)tbl <- r %>% MakeFrame() %>% tbl_df() %>% group_by(rho, Time) %>% summarise(power = mean(t))ggplot(tbl) + geom_line(aes(x = rho, y = power, col = factor(Time)))
library(dplyr)library(ggplot2)tbl <- r %>% MakeFrame() %>% tbl_df() %>% group_by(rho, Time) %>% summarise(power = mean(t))ggplot(tbl) + geom_line(aes(x = rho, y = power, col = factor(Time)))4. The MonteCarlo Package
Example: Power of DF-Test — ctd.
4. The MonteCarlo Package
Example: Power of DF-Test — ctd.
Conversion of the results into a LATEX table is particularly useful.
 # generate LaTeX table from simulation resultsMakeTable(r, rows = "Time", "rho")
# generate LaTeX table from simulation resultsMakeTable(r, rows = "Time", "rho")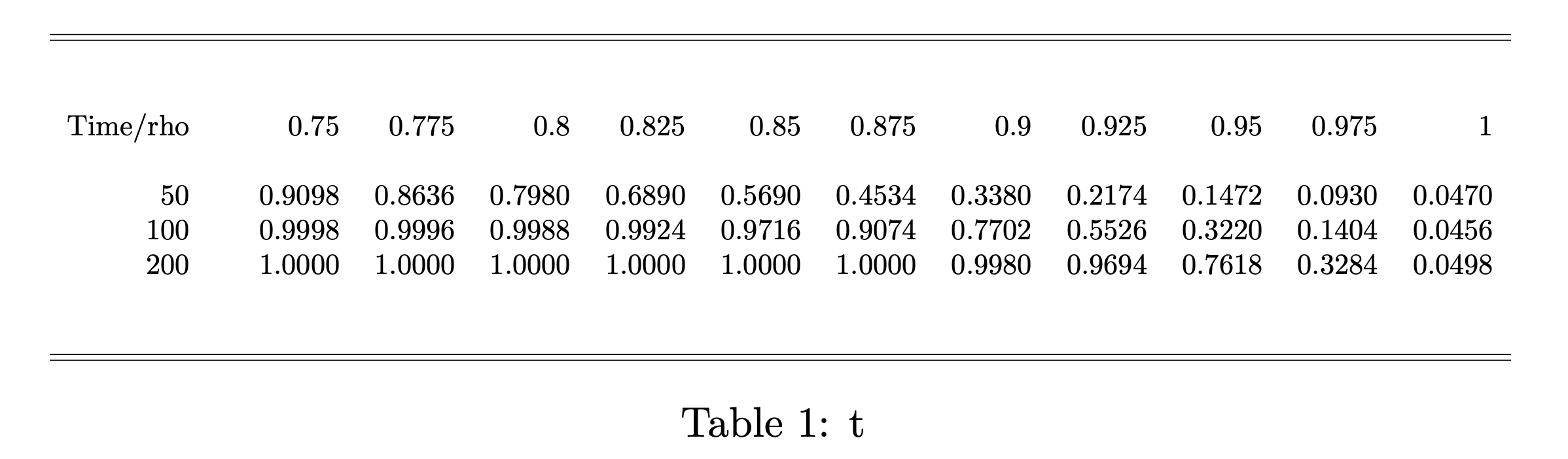
References
Gillespie, C. and Lovelace (2016), R. Efficient R Programming. O'Reilly Media.
Peng, Roger D. (2016). R Programming for Data Science. The bookdown Archive.
Wickham, H. (2019). Advanced R. 2nd Edition. Taylor & Francis, CRC Press.